Many misconceptions can creep into an undergraduate student’s mind as they study gene expression in their classrooms. In continuation with an earlier discussion on the common misconceptions in gene expression, in this article, educator Maya Murdeshwar from St. Xavier’s College, Mumbai highlights some of the misconceptions around the process of translation – the process of building a polypeptide chain based on genetic information.
Deoxyribonucleic acid (DNA) is the genetic blueprint in all living organisms on earth. It serves as the ‘instruction manual’ to create new life. The information stored within the DNA, in regions termed ‘genes’, is expressed transiently in the form of messenger ribonucleic acids (mRNA), and then as proteins. The change in biological alphabet from that of nucleic acids (in DNA and RNA) to that of amino acids (in proteins), is termed ‘translation’, akin to translating one language into another. While fascinating mechanisms weave together this process of ‘gene expression’[1], students are often confounded by the intricacies. In this second article around common misconceptions in gene expression, we will deal with two such misconceptions related to mRNA translation and protein synthesis.
Misconception 1:
‘The entire messenger RNA (mRNA) is translated into a protein’.
I realized this problem when I asked students to diagrammatically explain the process of translation starting with an mRNA sequence. Most students depicted the entire mRNA to be protein coding, failing to indicate the untranslated, non-coding regulatory regions.
This misconception arises when students fail to link mRNA structure to its translation – concepts that get covered in separate chapters in most textbooks.
Correct Concept:
During protein synthesis, translation begins at the ‘start’ or ‘initiation’ codon (usually AUG, rarely CUG and GUG) and ends at the ‘stop’ or ‘termination’ codon (either one of the triplets UAG, UAA, UGA).[1] The region between the initiation and termination codons is termed the ‘coding region’ that codes for the specific sequence of amino acids in a protein. However, the mRNA is much longer than the coding region.
A short stretch of untranslated nucleotides is present upstream (immediately preceding) of the start codon. Similarly, a short stretch downstream (immediately succeeding) of the stop codon does not encode amino acids and remains untranslated (Figure 1). These regions, present in both prokaryotes and eukaryotes, are termed the 5’-untranslated region (5’-UTR) or ‘leader’, and the 3’-untranslated region (3’-UTR) or ‘trailer’, respectively. These play a role in mRNA stability by providing sites for addition of the 5‑methyl guanosine cap at the 5’ end and the poly‑A tail at the 3’ end. The 5′- UTR is also where a ribosome – the site of protein synthesis in the cell – can bind to the mRNA and initiate translation.[1,2]
Further, in eukaryotes, the coding regions of mRNA (‘exons’) are interrupted by non-coding regions (‘intron’). The introns must be cleaved and removed and all the exons joined together (‘spliced) to give the complete, uninterrupted coding sequence in its entirety. This is achieved by a process termed ‘splicing’.[2]
While discussing translation, I recapitulate mRNA structure shown in Figure 1 so that students relate to the region being translated and those which are not.

Misconception 2:
Mixing up the orientations of codon-anticodon pairings while writing them in standard convention.
This confusion reveals itself when students are asked to recognize the correct codon-anticodon pairing from several given options (Figure 2a). Students often end up selecting an incorrect pairing as they are confused by the anti-parallel nature of the codon-anticodon interaction, and the necessity of representing all nucleic acid sequences in the 5’-3’ orientation (reading from left to right) as per standard convention.
Correct Concept:
During protein synthesis, the specific interaction between an mRNA codon and a tRNA anticodon specifies the amino acid that will be added to the growing protein chain. The mRNA is ‘read’ in the 5’ to 3’ direction, one codon at a time, by tRNA molecules carrying their cognate amino acids. The first two bases in the codon follow the rules of complementary base pairing with the corresponding bases in the anticodon (Figure 2b). However, more flexibility is accorded to the base in the third position of the codon, where non-complementary base pairing is tolerated. This is termed the ‘Wobble hypothesis’ and the third nucleotide in the mRNA codon, corresponding to the first in the tRNA anticodon, is called the ‘wobble base’.
The wobble base at the 3’ end of the mRNA codon (Figure 2b, red colour) thus pairs with the nucleotide present at the 5’ end of the anticodon, a 3’↔5’ correspondence in the pictorial depiction. However, while writing the anticodon sequence, the usual 5’-3’ convention is followed, creating confusion. For instance, consider the nucleotide sequences shown in Figure 2c. While codon 5’-CUU‑3’ pairs with anticodon 3’-GAA‑5’, the anticodon sequence is written as 5’-AAG‑3’ in the standard convention. Students are perplexed as to whether GAA or AAG pairs with the codon.
Moreover, if the sequence of an anticodon is provided and students are asked to determine the sequence of the corresponding codon and the amino acid it codes for, there is confusion galore! Consider the same example (Figure 2c). If the anticodon is 5’-AAG‑3’, the corresponding codon would be 3’-UUC‑5’, viz, 5’-CUU‑3’ as per the standard convention. A student confusing the 5’-3’ orientation might incorrectly infer from the standard genetic code[3] the codon to be UUC coding for amino acid phenylalanine, instead of CUU coding for leucine, which is the correct answer.
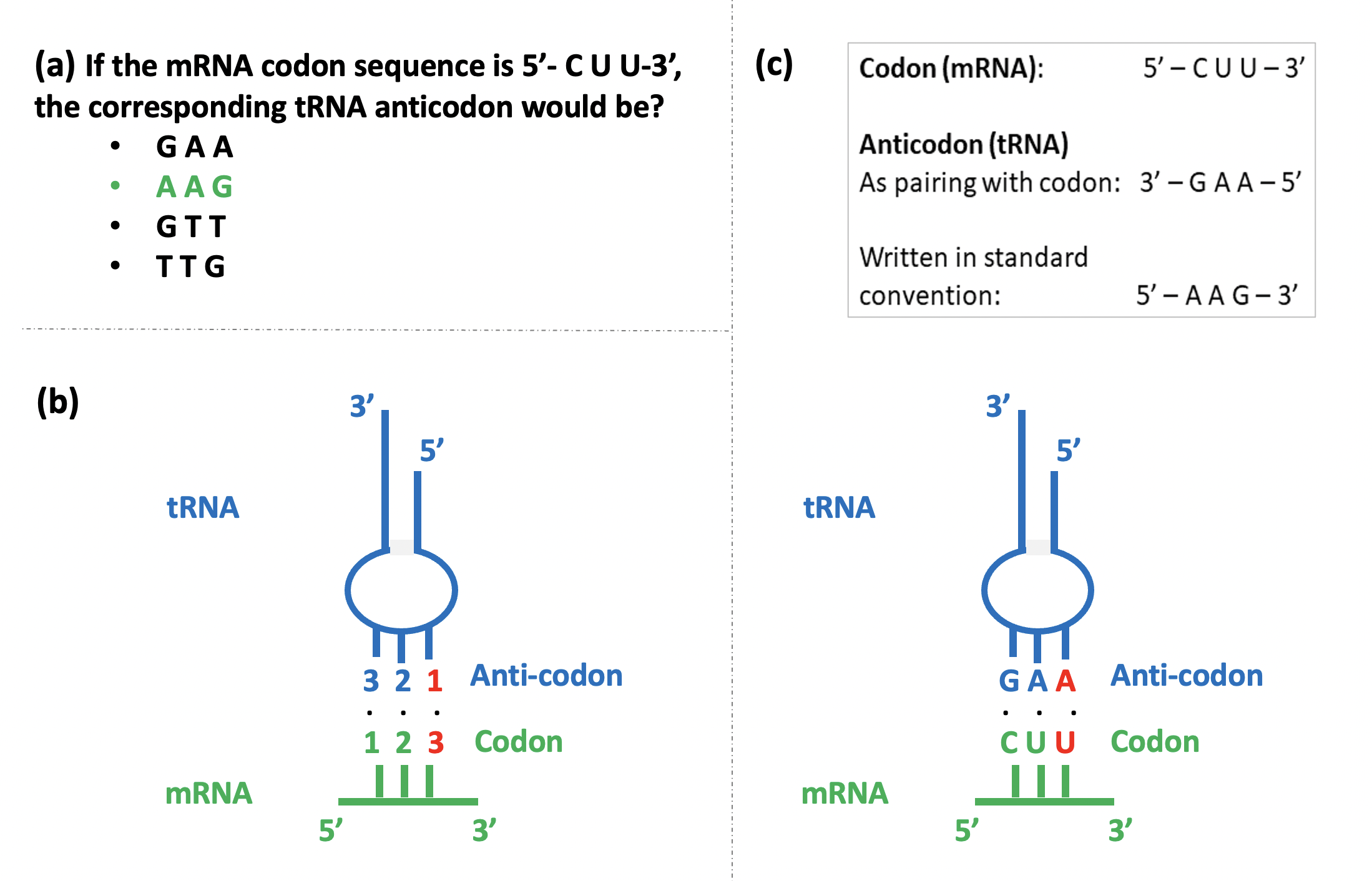
I usually clear this confusion by pictorially representing the codon-anticodon base pairing on the blackboard, with numbers and actual nucleotide sequences to represent the codon and anticodon, and by highlighting the orientation of the strands (Figure 2b and 2c). Then I invoke the standard convention of writing nucleotide sequences, and turn the anticodon sequence around in the 5’-3’ direction as shown in Figure 2c. I test their understanding with practice problems (Figure 2a) and then by reversing the question itself – by giving them an anticodon sequence (5’-3’) asking for the corresponding codon. Teachers could devise their own problem sets catering to the common and specific problems encountered in their classrooms.
It is immensely satisfying to experience the joy of students correctly grasping a concept – that ‘Aha!’ moment when their eyes brighten with the light of understanding and a happy grin spreads on their face.